-
Publish Your Research/Review Articles in our High Quality Journal for just USD $99*+Taxes( *T&C Apply)
Offer Ends On
Rosanna Capparelli and Domenico Iannelli*
Corresponding Author: Domenico Iannelli, Department of Agricultural Sciences, University of Naples Federico II, Via Università 100, 80055, Portici, Napoli, Italy.
Received: June 01, 2023 ; Revised: June 10, 2023 ; Accepted: June 13, 2023 ; Available Online: June 16, 2023
Citation: Capparelli R & Iannelli D. (2023) Introduction to the Study of the Microbiota. J Pharm Sci Drug Discov, 2(1): 1-8.
Copyrights: ©2023 Capparelli R & Iannelli D. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Views & Citations
Likes & Shares
The human gut includes about 1014 bacteria. Two microbial communities, Bacteroides and Firmicutes, prevail in the human microbiota, which displays high variability at the level of species and strains. This profile expresses the long-time coevolution between bacteria and host, which favored the early successful bacteria to prevail in diversity and evolve in species and strains. The gut microbiota has also a dynamic composition. Some bacteria persist throughout the live of the host, while others - originating from food, water, or other sources - are “transient”. Comparison of gene expression profiles from germ-free mice mono-colonized with Bacteroides thetaiotaomicron and non-colonized enabled to identify some genes involved in establishing a mutualistic colonization. This study clearly demonstrated how a single commensal organism could restore the multiple defects of a germ-free host. Mammalians and their microbiota coevolve towards mutualism. The long coevolution between host and microbiota explains how the host immune system could prevent commensals from leveraging host resources and maintaining immune tolerance to harmless bacteria. Understanding how beneficial bacteria influence the development of the host immune system may lead to natural therapies based on immuno-modulatory properties of microbiota.
Keywords: Microbiota, Phages, Immune system, Mutualism
INTRODUCTION
Microbial communities (or microbiota) are present in many environments, including water and soil. Through metabolic reactions, these communities built interspecies networks and a vital environment for the community. The microbiota of the human gastrointestinal tract (GI) - one of the many mutualistic communities - contains three domains of life: bacteria, archaea, and eukaria [1]. In this review, we treat only bacteria and bacteriophages (phages), the two most numerous communities in the GI [2]. GI microbiota may causes diseases, but more often develops conditions of mutual benefits for the host and the bacterial community [3]. We can say that our health depends much from the GI microbiota. Understanding the factors that confer stability to the GI microbiota of adult healthy subjects might help discovering how external factors - like bacterial infections - alter it, leading to microbiota-associated diseases. Our GI microbiota has about 100 times the number of our genes and provides us with genes and metabolites of its own [4]. As already mentioned, the host-microbiota relationship generally is mutualistic (both partners gain advantages from cooperating). Anaerobic bacteria degrade plant polysaccharides, that we ingest but cannot break down to monosaccharides and finally into short-chain fatty acids. From this mutualistic relationship, the host gains energy while bacteria gain a protected anoxic environment and abundant food (especially glycans, compounds consisting of a large number of linked monosaccharides). Microbial communities, whose integrated activity is a cost for the host, become target of the host selection and eliminated. Instead, microbial communities promoting host fitness expand their presence. Thus, the microbial diversity present in the GI reflects the selection for specific bacteria whose collective activity is beneficial to the host. This strategy promotes cooperation, diversity and stability of the microbiota, as confirmed by the remarkably stable and highly diversified microbiota of adult healthy subjects [5].
Phages are viruses that infect specific bacterial species and replicate by a lytic or lysogenic cycle [6]. In the lytic cycle, the phage genome colonizes the bacterium, replicates and the phage particles lyse the bacterium; in the lysogenic cycle, the genome of the phage integrates in the bacterial genome without killing the bacterium [6]. As with bacteria, phages may cause diseases, and - at the same time 0 prevent them. They contribute to the inflammation associated with obesity or type 2 diabetes [7] and can translocate across the mucosal barrier, reach the blood of healthy individuals and alter their immune system [8]. Prophage induction (the prophage is the bacteriophage genome integrated into the circular bacterial chromosome or existing as an extra chromosome or plasmid) may also contribute to intestinal dysbiosis changing symbionts to pathobionts [9], while antibiotics may induce production of phages from lysogenic bacterial strains [10]. As anticipated, phages also prevent diseases. They control the growth of the bacterial population, its diversity and metabolism [4]. In the first month of life, the microbiota of the infant expresses a large number of Caudovirals viruses [11]. At the age of two years – when infants change diet - prevail the Microviride. These results suggest that phages are essential for the health of infants [11]. Phages do not regulate only the bacterial population of the gut. They are present in about 45% of ascetic fluids from patients with bacterial infections. These data are particularly interesting since demonstrate that phages can translocate from the gut to other organs, opening the way to their potential applications in diseases where inflammation plays an important role, as in inflammatory bowel disease [12].
INTERACTIONS BETWEEN PHAGES AND BACTERIA
Phages have bacteria as their more frequent prey. Consequently, bacteria have developed numerous - and sometimes highly sophisticated - methods of defense, collectively referred to as “prokaryotic immune system” [13]. The bacterial antiphage defense genes generally reside in clusters or “defense islands”. To identify new defense tools against phages were studied the genes of unknown function residing near defense islands. The study led to the discovery of nine new families of antiphage defense genes and one family of antiplasmids, which protect against foreign DNA invasion [13]. In addition, the same authors found that the antiphage bacterial genes all have the Toll-interleukin receptor (TIR) domain. This study is particularly valuable: first, describes an ancient component of innate immunity (TIR), common to animals, plants, and bacteria; second, discovers thousands of systems that bacteria can use against phages. In addition, the discovery of the CRISPR-Cas anti-phage defense system [14] rapidly led to biotechnological and biomedical applications. Accordingly, it is plausible to anticipate that new systems (as the ones described in this and the next paragraph) will lead to a better understanding and manipulation of the human microbiota.
A CHEMICAL DEFENSE AGAINST PHAGE INFECTION
Phages are the main threat to bacteria. Proteins or protein-RNA complexes mediate all the anti-phage defense mechanisms so far described. Recently, Kronheim [15] described very small bacterial molecules - known as secondary metabolites (SMs) - which inhibit phage replication of competitors present in their niches [16,17]. Given the widespread role of SMs against cellular predators, Kronheim [15] screened 4,960 SMs (natural products, drugs and known bioactive molecules) to find SMs protecting E. coli from lysis by the phage λ. Bacteria grown for one hour in the presence of each SM and for five hours in the presence of the phage. In the absence of a SM, all bacteria died within two hours from phage addition. Instead, bacterial growth occurred in the presence of each of the 11 different SMs, indicating that the SMs inhibit phage replication. Nine out of the 11 SMs tested, resulted to be a DNA-intercalating agent and four more compounds resulted to be anti-cancer drugs. All secondary metabolites were isolated from Streptomyces, which produce a very large number of secondary metabolites [18]. Bacterial anti-phage defense systems previously known, all rely on protein components to mediate resistance. Here Kronheim et al. [15] demonstrate that Streptomyces species produce chemicals - other than proteins - with anti-phage defense system. Further - in contrast to other bacterial anti-phage systems active against a limited number of phages – SMs provide broad protection against all dsDNA phages. Further, being diffusible, a few DMs can protect the whole community and - since most bacterial species produce DMs - this anti-phage defense system is likely to be widespread in nature. Furthermore, the Streptomyces genus produces more than 100,000 antimicrobial compounds [19] most of which are as yet uncharacterized. Therefore, we expect that other classes of anti-phage molecules might exist. Identification of these compounds will reveal new groups of bioactive molecules that may have broad application for therapeutic purposes. Further investigations to identify and characterize these metabolites and their mechanisms of action will uncover new chemicals, expand our knowledge of bacterial anti-phage defense systems and improve our understanding of the evolutionary forces that shape bacterial communities.
HOST - MICROBE INTERACTION MOLDS THE GUT MICROBIOTA
The human gut includes about 1034 bacteria [20]. For long time, the structure of the human gut relayed on cultural methods. At present, are available molecular methods that consent to know the sequence of the bacterial 16s rRNA genes [21] and to study the human gut microbiota diversity, composition, and how the host-bacteria mutualism was born [22]. Two microbial communities, Bacteroides and Firmicutes, prevail in the human microbiota. In spite of the reduced diversity at the level of family, the human gut microbiota displays a high variability at the level of species and strains. This profile reflects the long-time of coevolution between bacteria and host, which favored the early successful bacteria to prevail on diversity and evolve into species and strains.
The gut microbiota has also the property of having a dynamic composition. Some bacteria can persist throughout the life of the host, while others - originating from food, water or other sources - are “transient”. At present, we know only in part how the resident bacterial species become permanent in a variable and competitive context. Comparison of gene expression profiles from germ-free mice mono-colonized with Bacteroides thetaiotaomicron and non-colonized enabled to identify several genes involved to establish a mutualistic colonization. Following colonization with Bacteroides thetaiotaomicron (B. thetaiotaomicron), the upregulated host genes include numerous genes participating to the cutaneous barrier function. Other upregulated host genes colonized with B. thetaiotaomicron, regulate postnatal maturation or angiogenesis. This study clearly demonstrates how a single commensal organism may restore multiple defects of a germ-free host. The ability of the B. thetaiotaomicron to recover energy from nutrients that otherwise would be indigestible for the host represents an ingenious solution of the bacterium to remain part of the host microbiota. The ability of this bacterium of feeding from different glycans confers stability to the microbiota providing polysaccharides to the community when dietary polysaccharides are scarce. This highly adaptive human gut symbiont - assuring the nutriment to the community - warrants also a permanent association with its host.
THE MUCOSAL IMMUNE SYSTEM DEPENDS ON THE COMMENSAL MICROBIOTA
The systemic human immune system evolved to keep the human body free from microbes. The mucosal immune system of the human gastrointestinal tract instead fosters a trillion of microbes that must live in a mutual relationship with the host and the intestinal microbiota. While the systemic immune system reacts almost automatically against any microbe it detects, the mucosal immune system must preserve its partnership with gut bacteria. Thus, the main property of the mucosal system is to tolerate the gut microbiota when the risk of infection is low and to respond with a tolerable immune response in case of disease or infection. When there is a low threat of infection, the microbiota avoids initiating an inflammatory response and preserve the cooperation with the host. The human host and the intestinal microbiota also establish a cooperative system, which stays alive only as long as the individual costs for maintaining the collaboration are lower than the benefits. Both partners work to maintain tolerance and thus both benefit of the mutualistic partnership. However, the collaboration terminates when the costs of maintaining the cooperative system are greater than the benefits.
Mammals and their microbiota co-evolved towards mutualism [22]. This very long and close relationship explains how the host immunity can prevent commensals from leveraging host resources and maintaining immune tolerance to harmless bacteria [23,24]. However, in a genetically predisposed host, imbalances between microbiota and immunity contribute to the pathogenesis of multiple immune-mediated disorders: circadian rhythm, nutrition, metabolism and immunity [25,26].
MICROBIOTA - IMMUNE RESPONSE INTERACTION
The function of specific T cell populations is associated with the presence of specific bacteria. The absence of specific microbes may alter the function of specific immune cells. Independent studies have shown that Th7 cells are rare in the small intestine of germ-free mice. The level of Th7 on mucosal T cells depends from the presence of segmented filamentous bacteria (SFBs) adhering to the epithelium of the small intestine. Mice normally colonized - but lacking SFBs - show a low number of Th7 cells in the small intestine, as germ-free animals. Upon colonization with SFBs, the germ-free mice display levels of Th17 comparable to those of control animals. Further, treatment of SFBs-colonized cells with antibiotics kills the microbiota and cause the loss of Th17 cells [27,28].
In another study, the polysaccharide A (PSA) expressed by the Bacteroides fragilis modulates both the mucosal and systemic immune systems. Mono-colonization of CD4+ T cells with B. fragilis induces IL-10 production from these cells, which in turn protect against colitis [29]. However, once the B. fragilis infection is systemic a pro-inflammatory response leads to IL17-dependent peritoneal abscess [30]. Abscess formation provides clear evidence that the mucosal immune system can reverse between toleragenic and inflammatory response to the same microbe [31]. Additionally, one could suppose that once the risk of systemic infection by B. fragilis is finished, PSA might once again induce IL-10 production from CD4+ cells among mucosal T cells, to help the mucosal immune system to switch back from inflammation to cooperation with the microbiota. The ability of antigen-specific T cells to switch between pro- and anti-inflammatory subtypes is also a plausible hypothesis of how the mucosal immune system can shuffle between the inflammatory and toleragenic immune responses necessary to maintain intestinal homeostasis. Alternatively, the microbiota may also direct T cells to adopt the immune profile necessary for maintaining homeostasis or suppose that loss of the host or the microbial components might serve to facilitate the switch from inflammation to immune disorders such as IB.
THE MICROBIOTA BETWEEN HEALTH AND DISEASE
The evidence that intestinal bacteria interact with the immune system of the host shows the importance of the microbiota composition for the host health. If specific classes of bacteria promote the host’s health, then diseases may derive from the absence of these bacteria and their critical molecules. Inflammation emerging from dysbiosis between symbionts and pathobionts [29] leads us to think that many similar intestinal diseases may exist and to ponder why the study of probiotics (bacteria such as lactobacilli promoting the health of the host) so far did not identify bacterial molecules protecting the host [29]. The evidence that a single molecule can control inflammatory diseases suggests that many other bacterial molecules promoting human health wait for discovery. The human symbiont Bacterioides fragilis protects animals from experimental colitis induced by Helicobacter hepaticus, a prospective pathogenic commensal bacterium. This effort needs a single microbial molecule (polysaccharide A, PSA) [29].
In the near future, the finding that PSA from B. fragilis is a natural anti-inflammatory molecule may lead us to discover therapies based on the knowledge of the relationship between humans and their microbial partners.
HOST GENOME-MICROBIOTA INTERACTIONS
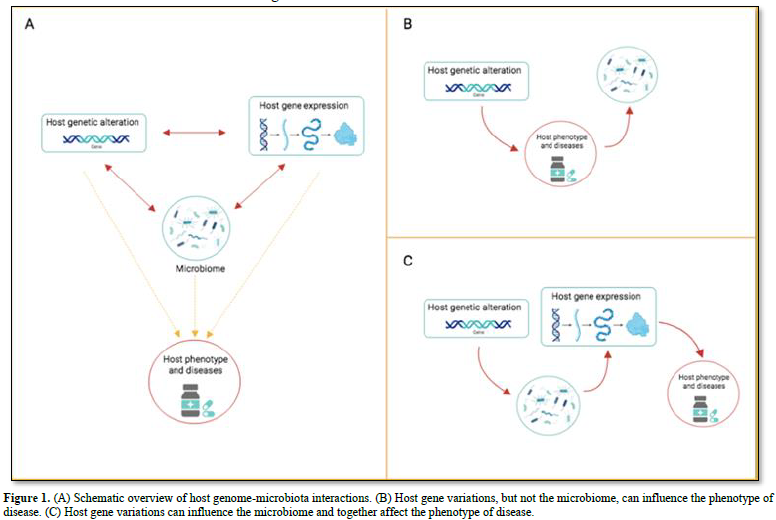
Several features of the host shape the human microbiota: genome, diet, age, sex, weight, and diseases (bowel disease, type 2 diabetes, colorectal cancer and several more) [32,33]. Many diseases are under the influence of both the microbiota as well as the host genes. Thus, it could be very informative knowing the individual contributions of the microbiota and of the genes on the disease. One of the first host gene-microbiota interaction concerns the genes FUT2 (encoding fucosyl transferase 2 and MEFV Mediterranean fever) [34]. Other studies concentrated on the heritability of the microbiota composition. The results from several hundred samples of gut microbiota from mono- and di- zygotic twin pairs showed that several bacterial taxa are hereditable. Studies using germ-free mice established that the Christensenellacae taxon is significantly heritable in obese hosts [35]. A more recent study detected that the inflammatory bowel disease (IBD) is associated with 49 microbiota gene loci, including innate immune response pathways [36]. The same study detected an association between the human gene LCT and abundance of Bifidobacterium in the gut microbiota. The LCT gene encoding the lactase gene enzyme (which metabolizes lactose), while the Bifidobacterium uses lactose as a carbon source. The field of the host genome – microbiota interactions is relatively recent. However, it is already clear that the association between the two components can have different directions of causality (Figure 1A). There are cases where the host genes (but not the microbiota) influences the phenotype of the disease (Figure 1B). In this case, changes in the microbiota are attributable to the disease, or to disease-related changes (inflammation or medicaments). For example, antidiabetic drugs alter gut microbiota results [37]. Since long time we known that gene variation can lead to change the phenotype of a disease; only recently we learnt that changes in the microbiota can have the same effect. The NOD2 gene offers an example. Mice lacking NOD2 are prone to colitis, which is transmissible to wild-type hosts via the microbiota [38]. A third scenario describes the interaction between host genetic variation and microbiota influencing host gene regulation (Figure 1C). In the colonic epithelium of germ-free mice – following colonization with fecal microbiota - have been detected genome-wide changes in gene expression, including down-regulation of genes, together with genes active in the transport and metabolism of lipids and several more nutrients [26]. The authors found that the host response to microbial colonization varies according to the intestinal region and time elapsed since colonization. An independent study found that microbiome colonization induces activation or inactivation of a large number of enhancers in the mouse colon [39]. These results demonstrate the importance of studying how the microbiota uses to control host gene regulation.
THE IMPORTANCE OF GLYCANS
The mutualistic relationship between host and intestinal Bacteroides is set up on glycans. The host provides plant polysaccharides and receives products of bacterial fermentation. Glycans from the bacteria are essential for the survival of the bacteria and because provide to the host immunomodulatory molecules. The human digestive tract includes the small intestine and the large intestine (colon). The two compartments are interrelated. Digestion starts in the mouth, where the starch - once broken down by the amylase - proceeds to the stomach, where the acid pH and enzymes break down the proteins. The acidic food and the partially digested food enter the small intestine, where the macromolecules (lipid, proteins, nucleic acids, and carbohydrates) are broken down, vitamins are absorbed, and lipids are degraded. The majority of the undigested macromolecules in the colon is dietary fibers. In the colon, predominate the Bacteroides, whose major role is the production of glycans. These bacteria produce a large variety of enzymes essential to short–chain fatty acids (SCFAs) that provide different types of glycans essential for bacterial survival and immunomodulatory molecules to the host [40]. The intestinal Bacterioides produce a large variety of glycosylated molecules. A single strain of Bacterioides fragilis synthetize eight distinct capsular polysaccharides [41], an extracellular [42] and a large number of glycoproteins [43]. The B. fragilis 9343 encodes about 80 glycosiltranspherase. The expression of each of the eight capsular polysaccharides of B. fragilis is subject to phase variation regulated by DNA inversion of the promoter, conferring great antigenic variability to these organisms [41]. The synthesis of multiple phase-variable capsular polysaccharides is a conserved feature of intestinal Bacterioides, but not to other closely related species, suggesting that this property is advantageous only to intestinal survival. The ability of these bacteria to express subpopulations of organisms expressing distinct capsular polysaccharide biosynthesis loci are heterogeneous within the species, meaning that different B. fragilis strains express distinct capsular polysaccharides.
THE INDUSTRIALIZED MICROBIOTA
People moving from a country with a “non-industrialized” microbiota to one with an “industrialized” microbiota experience alterations of their original microbiota. Compared to the “non-industrialized” microbiota, the “industrialized” one exhibits a lower microbial diversity [44]. Antibiotics, processed food, and a sanitized environment, all influence the microbiota. In the industrialized countries, these actions were adopted ignoring the effects they could have on the human microbiota. Consequently, the microbiota of people living in “industrialized” countries resulted having characteristics never seen before in humans. “Industrialized” microbiota results from recent progresses in medicine, diet, and sanitation.
Medicine has reduced our exposure to pathogenic microbes and technology nourished populations. Again, these advantages came at cost of damages inflicted to our microbiota, ignoring the importance of the gut microbiota for our health, and the strong relationship between composition of the gut microbiota and alteration in the immune system of the host. These relationships connect the “industrial” microbiota to the long list of chronic diseases caused by inflammation. Remarkably, researchers discovered that these diseases diffuse along with the factors that alter the microbiota. Once understood how the microbiota influences human health, rapidly it was evident that the global spread of the industrial lifestyle would cause the global spread of chronic diseases, an event plausibly not easy to remove [44].
Microbiota biodiversity conservation is essential to safeguard human health. The benefit of the services provided by the microbiota to humans has been compared with the benefit of ecosystem services to the macro ecosystems [44]. At present, it remains the obstacle of identifying the changes that industrialization induced to the microbiota and presumably to the human health.
FUTURE STUDIES: DEVELOPING TRANSLATIONAL POTENTIAL
Microbiota research is moving from laboratory observations to clinical studies. This effort requires collecting data on immune system and the microbiota - two structures strictly connected - from representative patient populations. These variables may lead to non-invasive and advanced diagnostic procedures. In the future, it may be possible that the surgeon - before the intervention - will request a sample of stool, vaginal or saliva samples (the saliva provides efficient microbial biomarkers also from organs different from the mouth) [45]. This additional information - along with data from genome sequence and the medical history of the patient - helps the team to reach a more accurate prediction on the intervention outcome. However, reaching this objective requires knowing how the microbiome of the patient varies during the health and the disease states.
The analysis of the microbiota provides also a more accurate stratification of the patients and new biomarkers of several diseases. The techniques necessary to characterize the microbiota and metabolism already exist, but the methods need to be standardized. Microbiota analysis is rapidly becoming routine into clinical investigation. At the same time, clinical statistics tends to integrate within the microbial statistics. This trend in turn will facilitate the integration of patient studies with animal studies enabling a more exact interpretation of the data. Briefly, in the next decade the patient care will improve significantly, while the study of the microbiota is leading to personalized medicine: to more effective diagnosis and better disease treatment. In addition, the reduced cost of cancer genome sequencing permits the rapid identification of the treatment leading to the more favorable outcome in the individual patient with a disease, such as with colorectal cancer [46].
CONCLUSION
Mammalians support their microbiota from millennia. Thus, the host- bacteria co-evolution had time to create a very complex mixture of microbiota and hosts [47]. Imbalances in the microbiota may contribute to several human diseases, and alter the conformation of the gut bacteria. There is clear evidence that B. fragilis protects its host from an inflammatory disease caused by H. hepaticus in an animal model of experimental colitis [29]. The evidence that intestinal bacteria interact with the host’s immune system highlights the importance of the microbiota composition for the host’s health. If specific classes of bacteria have indeed evolved to promote the host’s health, then disease may well originate from the absence of the organisms and their essential molecules (for example, as result of improved hygiene). Inflammation resulting from dysbiosis between symbionts and pathobionts may represent the molecular foundation for many intestinal – and plausibly also non-intestinal - diseases. The studies of probiotics so far have failed to identify specific bacterial molecules able to protect the host [48]. There is evidence that a single bacterial molecule can control an inflammatory disease in animals [29]. These results provide evidence that molecules that have evolved to promote human health exist and wait for their discovery. The finding that a single microbial molecule (polysaccharide A, PSA) from B. fragilis is a natural anti-inflammatory molecule that promote mammalian health may stimulate the development of therapy based on the relationship between humans and their microbiota.
No Files Found
Share Your Publication :